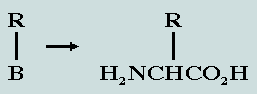Introduction
The work of shCherbak is particularly
apposite in this context. Predicated upon the nucleon counts for
distinct groupings of the 20 canonical amino acids through their
representation in the 64 codons of the standard model, his
criterion of ‘divisibility by 037’ is axiomatic to the
underlying organising principles inherent in the genetic code.
Many of the numerical features symmetrically depicted within the
Star of Israel matrix are dependent on this same criterion, i.e.
‘divisibility by 037’, as may be observed in Part 1 of
this series. Boulay, too, plays a vital part in these
proceedings, his proline hypothesis bringing the number of atoms
in the 64 codons up to a multiple of 37. Thus, in commencing our
analysis, a discussion, and resolution, of the ‘proline
problem’ is in order.</p>
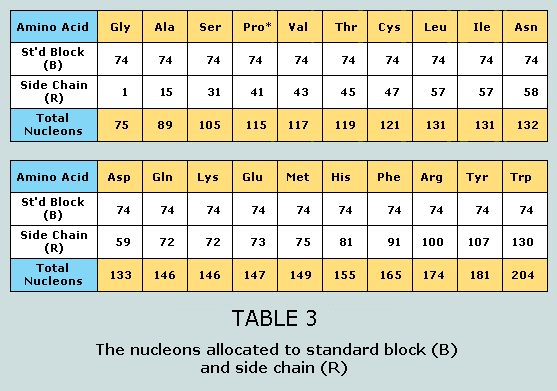
The proline problem It transpires that 19 of the 20 canonical
amino acids comply with the generic formula: Here, the lower component represents the
so-called standard block – designated B.
The variety in amino acid characteristics derives entirely from R
– the unique side chain. Thus, for 19 of the
canonical amino acids the nucleon count for the standard block is
74. The one exception to this general rule is proline,
which has one less hydrogen bonded to the nitrogen of its
standard block. In order to harmonise the nucleon counts for the
standard blocks of all 20 canonical amino acids, and thereby to
bring into play the extraordinary numerical phenomena that
eventuate, shCherbak suggests the moving of a hydrogen atom from
R to B – thus standardising both atom and nucleon counts for
B but decreasing those of R (respectively, 9 and 42) each by one.
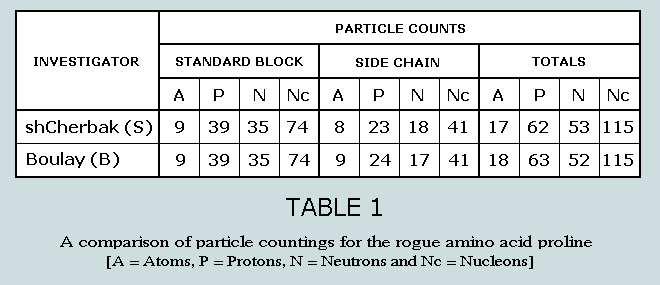
On the other hand, Boulay posits a neutron from R functioning as
a proton as it connects to the standard block; the result –
an extra proton (and thus a hydrogen atom) in B and one fewer
neutrons in R. There follows a summary of these proceedings: Boulay
defends the validity of this manoeuvre as follows: “Without
these two special countings, the very numerous and organised
phenomena presented by shCherbak and Boulay disappear. (But
they) are much too numerous and structured to be destroyed by
an apparent contradiction of chemical arrangement.” Number of atoms in the 64 codons As a result of Boulay’s proline
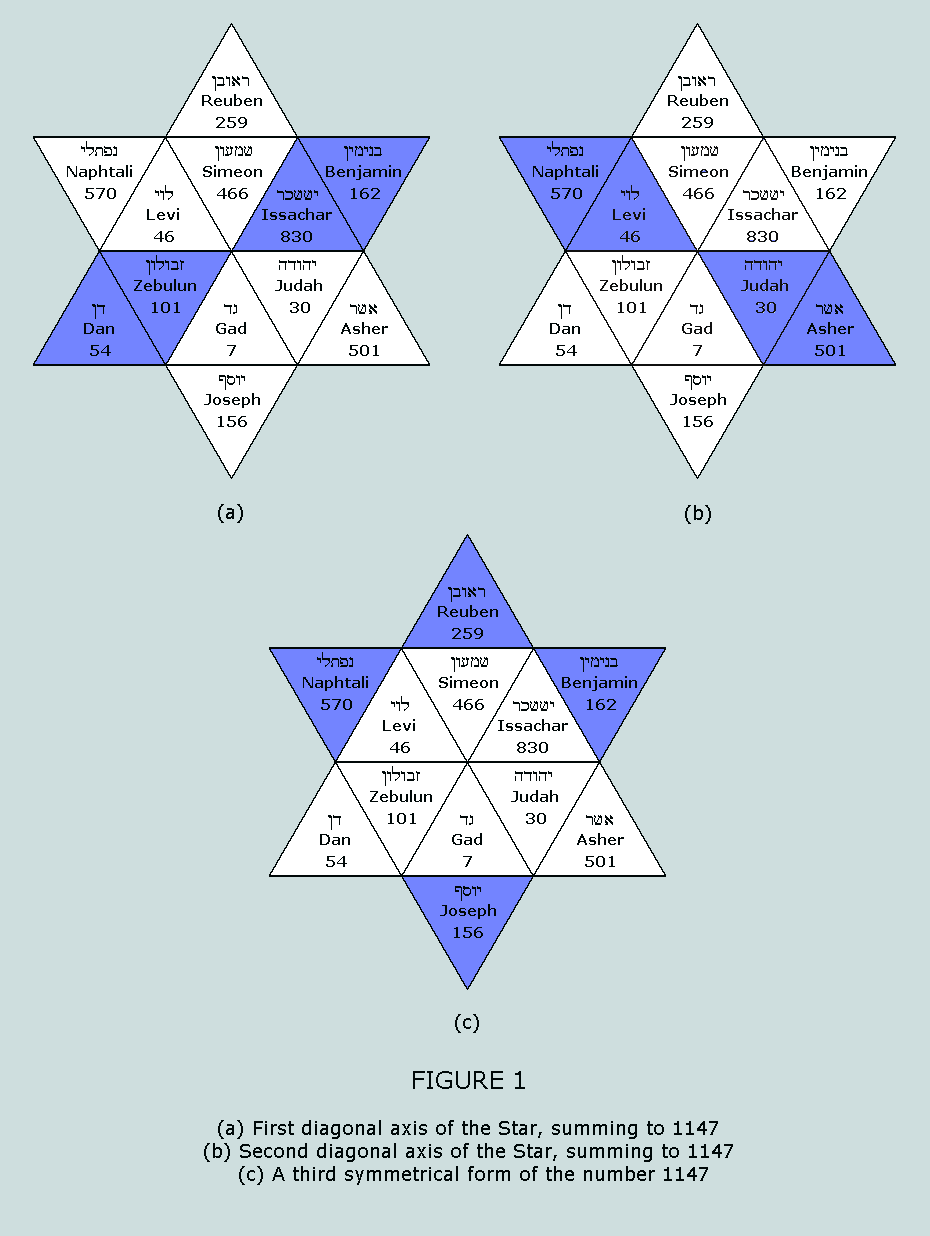
hypothesis, the following Table is prepared: It will be observed that the 64 codons
return a total atom count of 1147 = 37 x 31. This number occurs
symmetrically within the Star of Israel matrix on no less than
three occasions: Having observed the number of atoms in the
64 codon table of the amino acids to be 1147 – a number that
meets shCherbak’s axiom of ‘divisibility by 037’
– after the application of Boulay’s proline hypothesis,
and having observed the symmetrical composition of this same
number on no less than three occasions from within the Star of
Israel matrix, we now proceed to the standard blocks of the 20
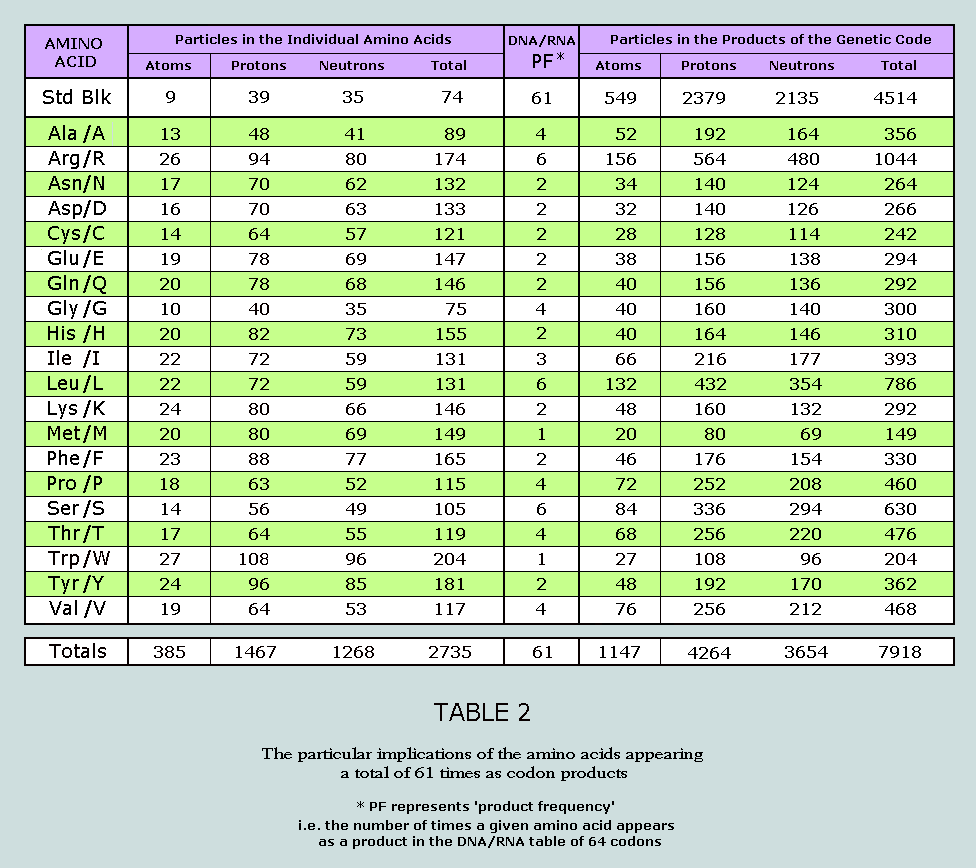
canonical amino acids. Nucleons in the standard blocks of the 20
canonical amino acids Following Boulay’s resolution of the
proline problem, the nucleons of the 20 canonical amino acids can
now be separated into those in the standard blocks and those in
the side chains, as per the following Table: It will be observed that the standard blocks
of the 20 canonical amino acids return a total nucleon count of
1480 = 37 x 40. This number occurs symmetrically within the Star
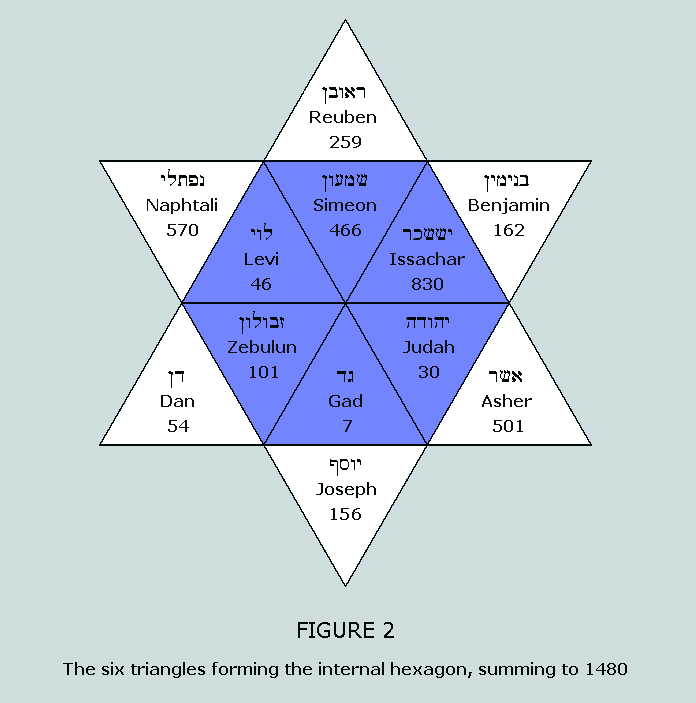
of Israel matrix in a most impressive manner: Having established the number of nucleons in
the bases of the 20 canonical amino acids to sum to 1480, and
having observed the symmetrical manner in which this same number
is arrayed within the Star of Israel matrix, we turn our
attention now to aspects concerning the nucleon counts for the
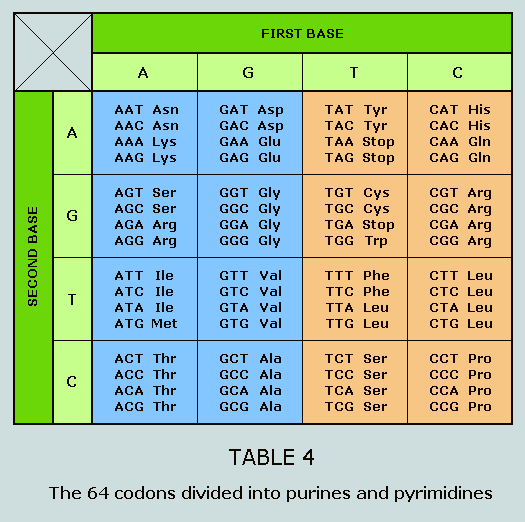
purines and the pyrimidines. The purine and pyrimidine codon groups It has been established that the 64 DNA
codons divide into two groups – those that begin with A or
G, and those that begin with C or T. These groups are referred to
as the purines (coloured blue in the following Table) and the
pyrimidines (coloured orange), respectively: Having thus made a division between the
purines and the pyrimidines, we now proceed to analyse their
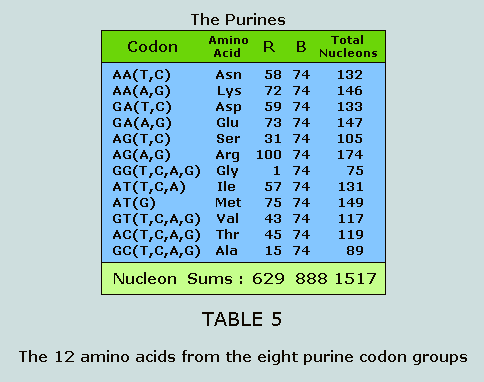
respective data, beginning with the purines. The 12 amino acids of the purine codon
groups The 32 purine codons break into eight groups
from which we extract 12 amino acids for analysis: These respective nucleon count numbers
appear in a compellingly symmetrical manner within the Star of
Israel matrix: These same numbers featured at Figure 3
appear in yet another symmetrical form within the Star of Israel
matrix: Having established the symmetry of the
nucleon counts for both the side chains and the standard blocks
of the purines within the Star of Israel matrix, we now proceed
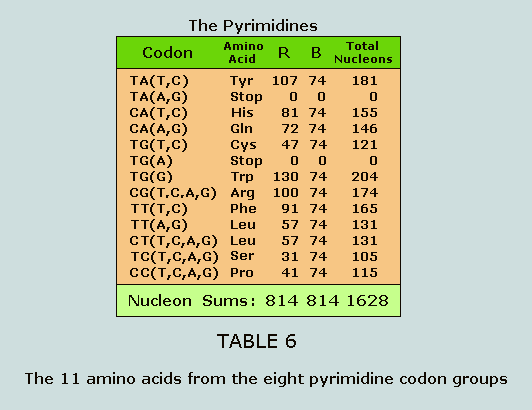
to the pyrimidines. The 11 amino acids of the pyrimidine
codon groups The 32 pyrimidine codons break into eight
groups from which we extract 11 amino acids for analysis: These respective nucleon count numbers,
being the same, appear symmetrically within the Star of Israel
matrix: Nucleons in the side chains of the
purines and the pyrimidines As observed at Table 5 and Table 6, the
nucleons in the side chains of the purines and the pyrimidines
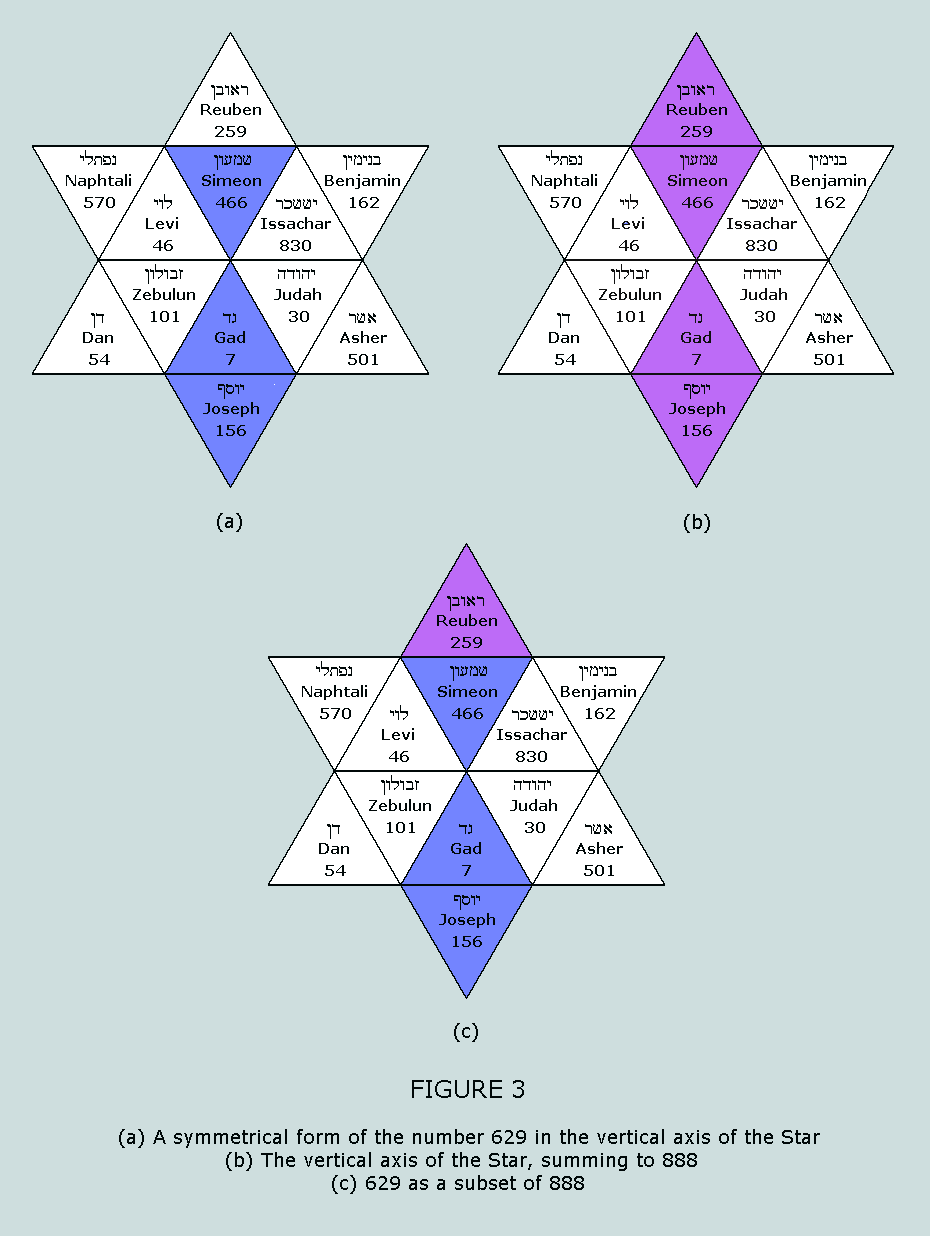
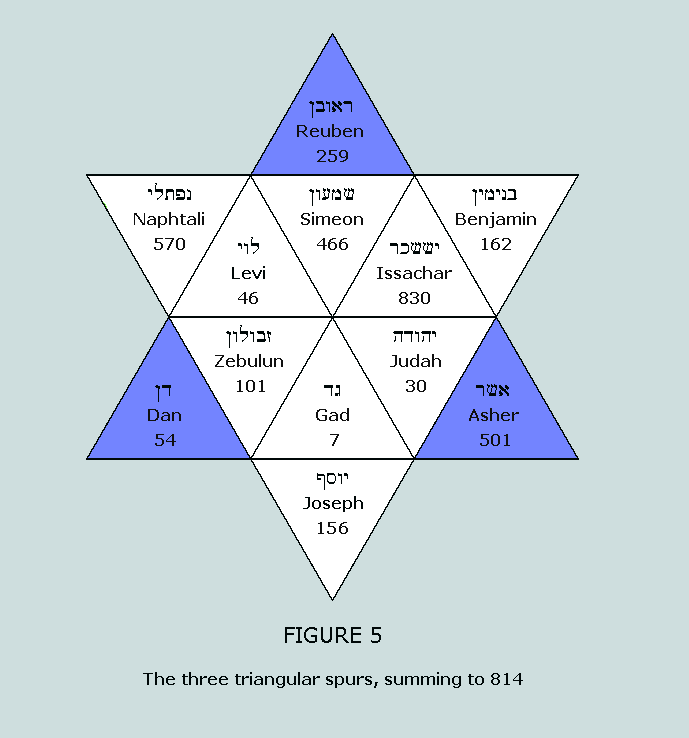
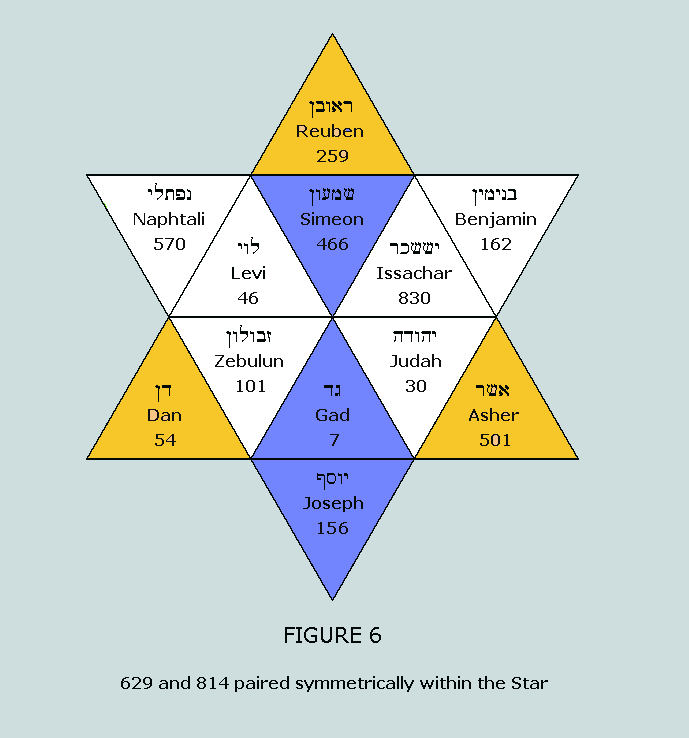
sum to 629 and 814, respectively. These numbers appear in a
fascinating symmetrical manner within the Star of Israel matrix: Nucleons in the standard blocks of the
purines and the pyrimidines From Table 5 and Table 6 we once more
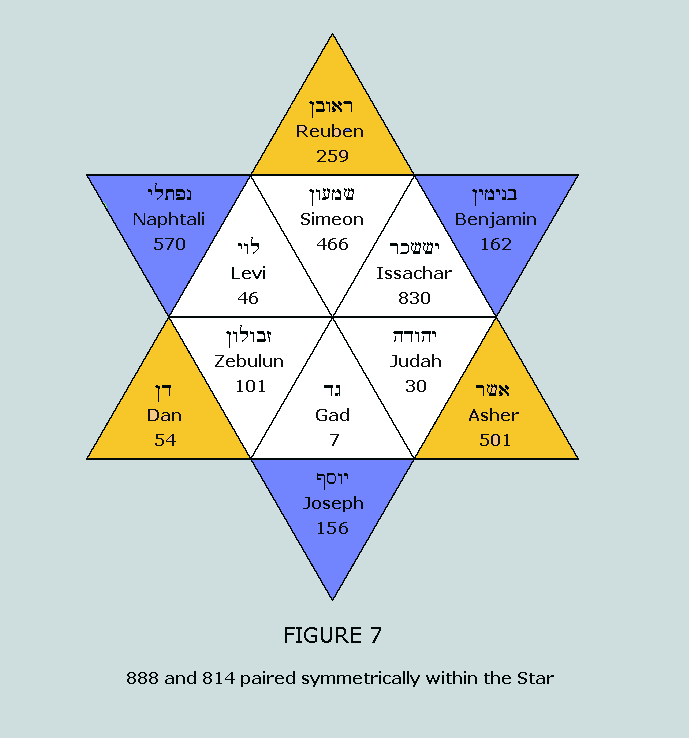
observe that the nucleons in the standard blocks of the purines
and the pyrimidines sum to 888 and 814, respectively. These
numbers appear in a most convincing fashion within the Star of
Israel matrix: The exquisite symmetry of this feature is
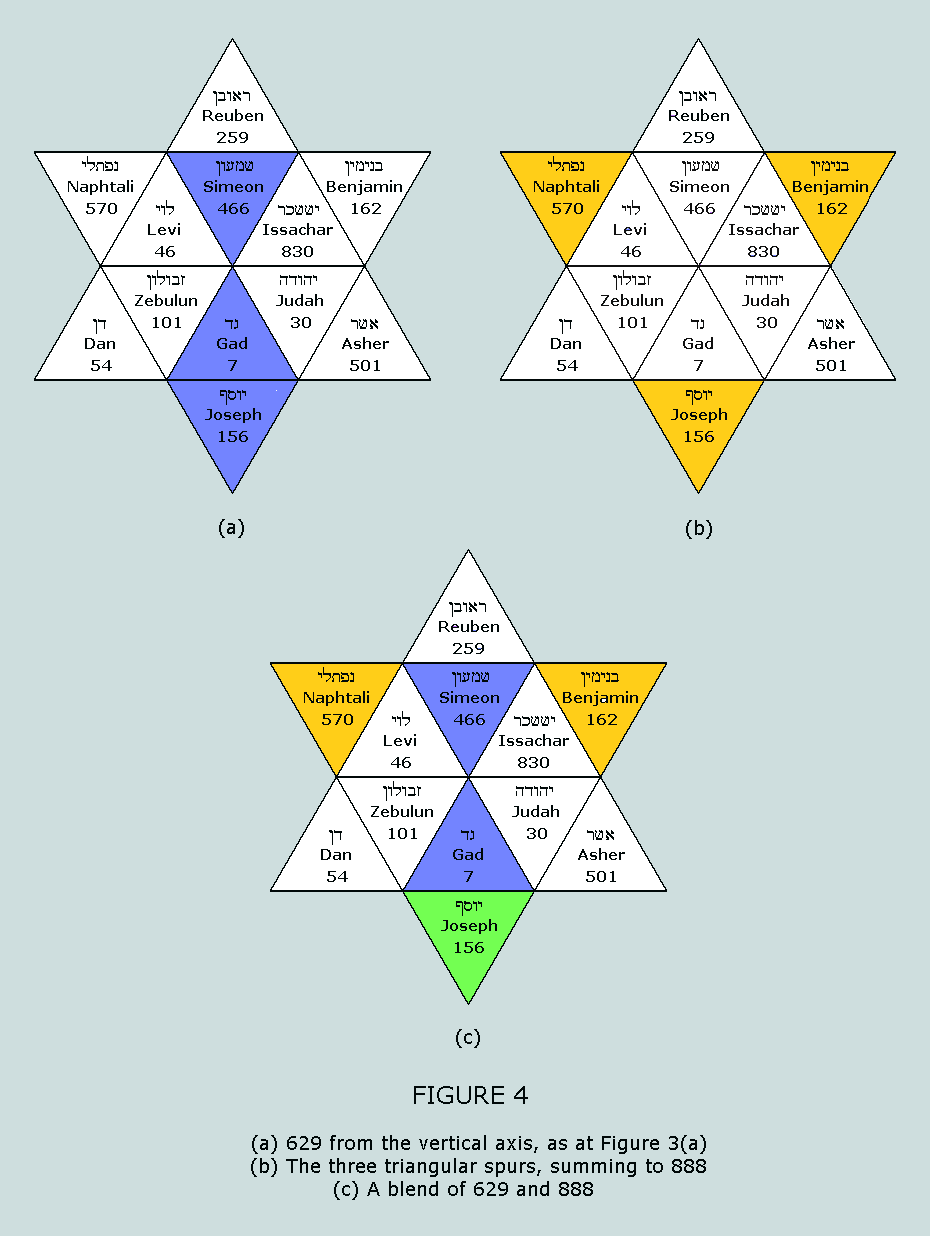
not its only symmetrical representation within the Star, as
evidenced by the following: The four CVs
forming the vertical axis sum to 888 = 37 x 24; these
represent the number of nucleons in the standard
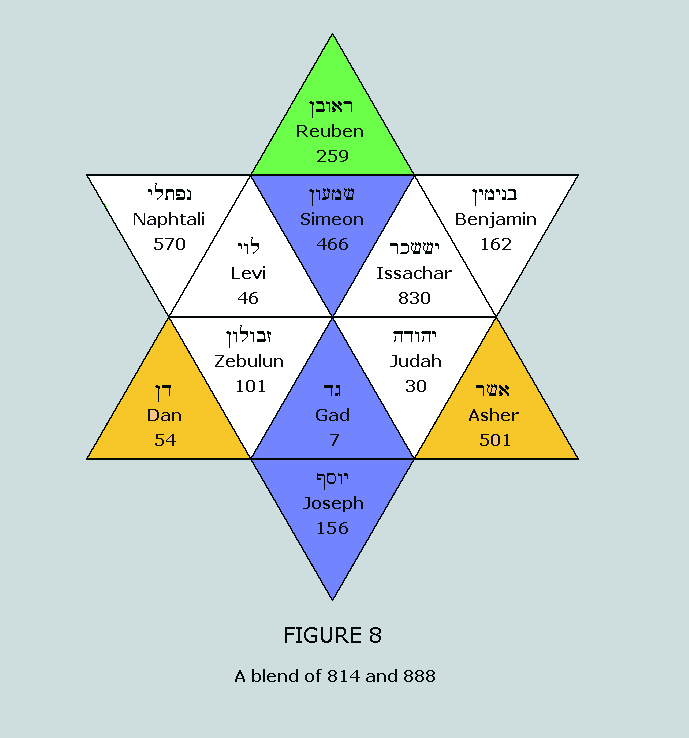
blocks of the purines Clearly, Figure 8 reworks Figure 6 into a
blended representation of 814 and 888. Therefore, the same six
CVs depicted in both Figures can be used to represent: (i)
The nucleon counts for the side chains of both the
purines and the pyrimidines – 629 and 814,
respectively (ii)
The nucleon counts for the standard blocks of both the
purines and the pyrimidines – 888 and 814,
respectively This feature exemplifies in astonishing
fashion the coincidences between some of the major patterns in
the genetic code found by shCherbak, and those pertaining to
symmetrical formations embodied within the Star of Israel matrix.
Both numerical patterns pertaining to the purines and the
pyrimidines at Figure 6 and Figure 8 – written into Nature
by their constituent nucleons, and into the CVs of the Star of
Israel by gematria - are dependent on shCherbak’s criterion
of ‘divisibility by 037’. Before closing this analysis, a few more
patterns are worth noting. These correspond to discrete
symmetrical arrangements of the 64 codon table and their
numerical analogues in the Star of Israel matrix, also
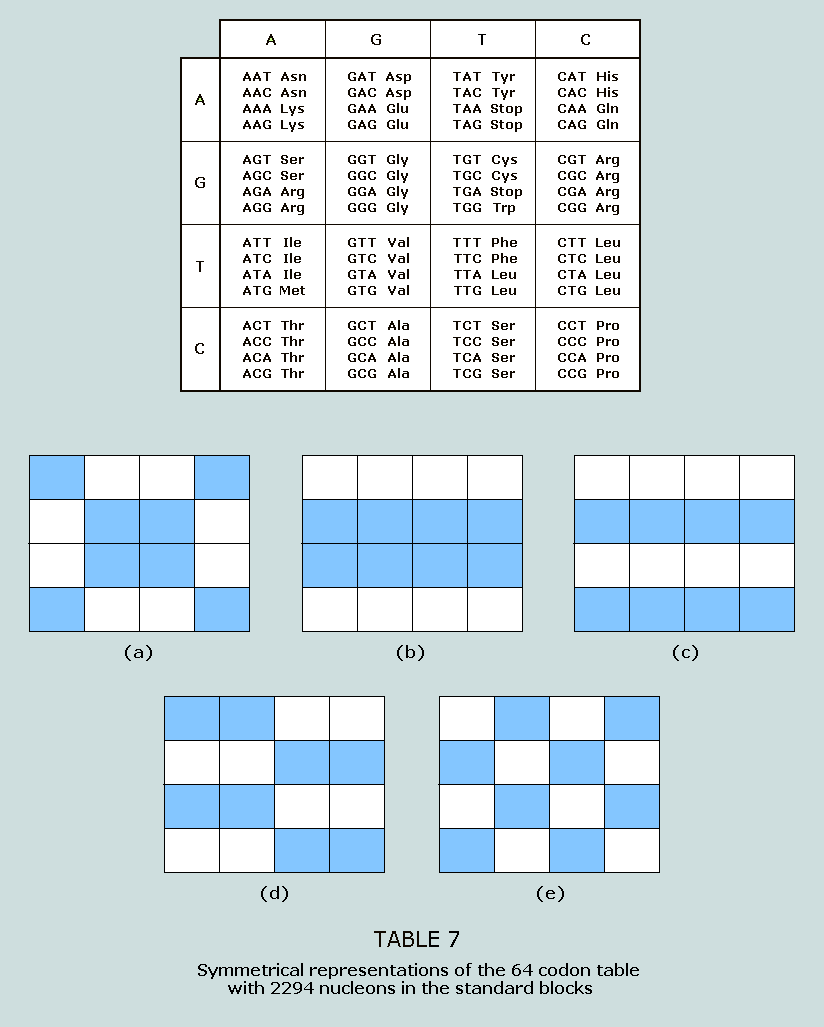
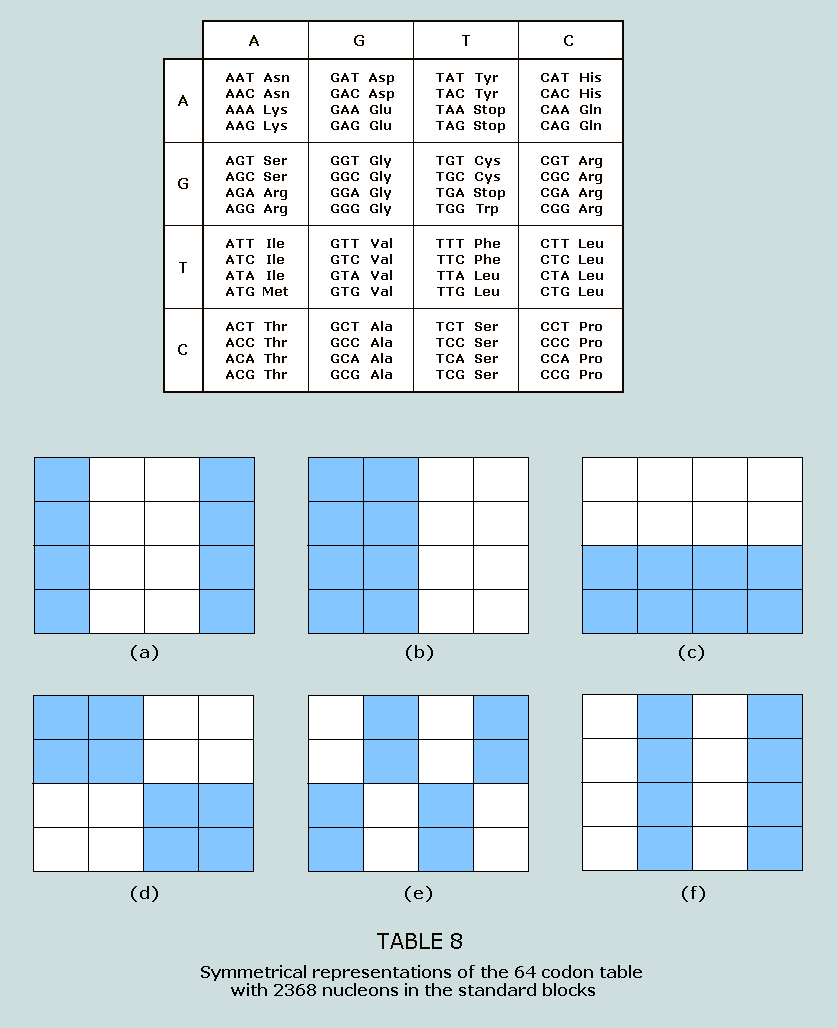
symmetrically disposed. 2294 nucleons in the standard blocks: The 64 codon table can be symmetrically
highlighted to produce arresting visuals, which follow: Table
7(a) consists of amino acids beginning with AA, CC, GG, TT, AC,
CA, GT, and TG, the number of nucleons in the standard blocks for
the 32 codons represented sums to 2294 = 37 x 62 Note
that one of the codons – TGA – is a stop codon Table
7(b) consists of amino acids with either G or T in medial
position; again, the number of nucleons in the standard blocks
for the 32 codons represented sums to 2294 = 37 x 62 Table
7(c) consists of amino acids with either G or C in medial
position; this is the same total as before Table
7(d) consists of amino acids beginning with AA, AT, GA, GT, TG,
TC, CG, and CC; the same total as before Table
7(e) consists of amino acids beginning with AG, AC, GA, GT, TG,
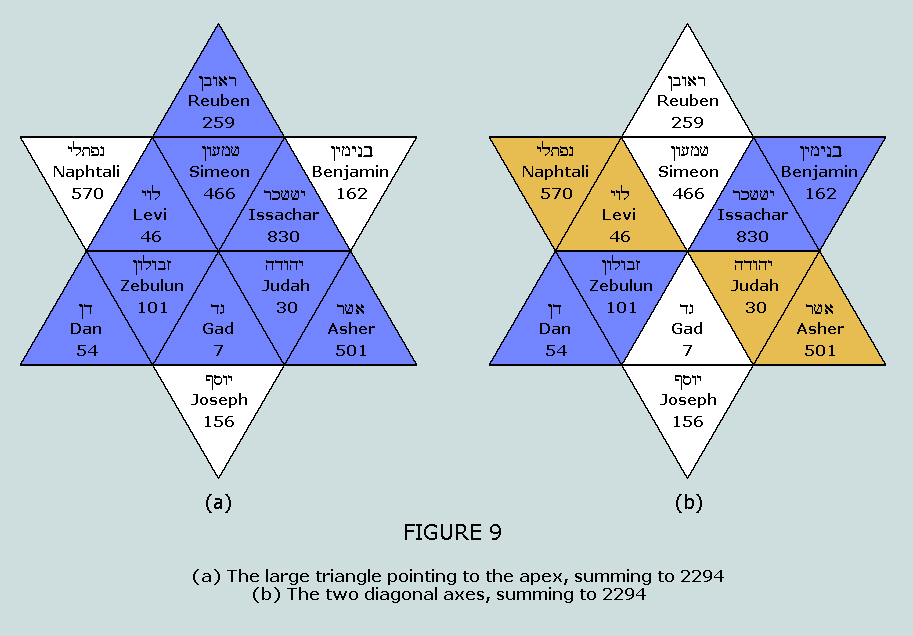
TC, CA, and CT; the total, as before 2294 within the Star of Israel The number 2294 appears symmetrically within
the Star on two occasions: The large triangle
at Figure 9(a) sums to 2294 = 37 x 62 The
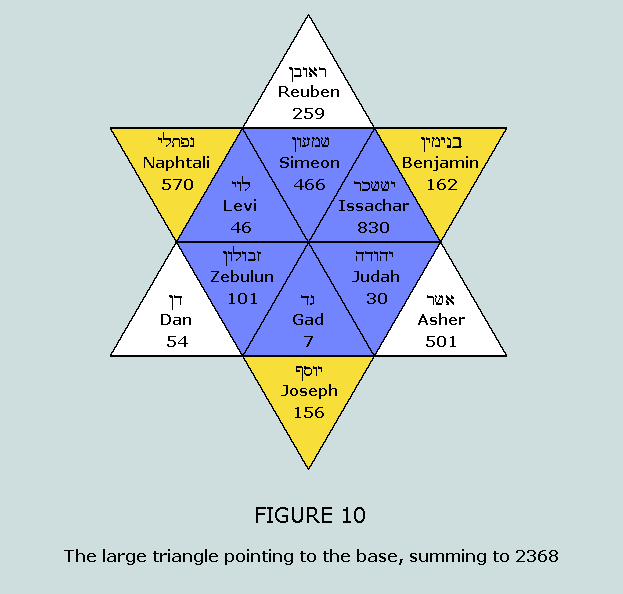
final patterns are now prepared for our analysis. 2368 nucleons in the standard blocks: The 64 codon table reveals another relevant
set of patterns by symmetry: Table
8(a) consists of amino acids beginning with either A or C; the
number of nucleons in the standard blocks for the 32 codons
represented sums to 2368 = 37 x 64; 2368 is the CV of the
Lord’s name, Jesus Christ (IhsouV
CristoV) Table
8(b) consists of amino acids constituting the purines; the number
of nucleons in the standard blocks of the 32 purine codons sums
to 2368 Table
8(c) consists of amino acids with either T or C in medial
position; the number of nucleons in the standard blocks for the
32 codons again sums to 2368 Table
8(d) consists of amino acids beginning with AA, GG, TT, CC, AG,
GA, CT, and TC; the number of nucleons in the standard blocks for
the 32 codons, 2368 Table
8(e) consists of amino acids beginning with AT, AC, GA, GG, TT,
TC, CA, and CG; the number of nucleons in the standard blocks for
the 32 codons, 2368 Table
8(f) consists of amino acids beginning with either G or C; the
number of nucleons in the standard blocks for the 32 codons, 2368 Summary By using the same criterion of
‘divisibility by 037’ featured so prominently in the
work of shCherbak, the Star of Israel has been observed to
reproduce numerical patterns previously found and described from
within the genetic code by the former. Furthermore, these
numerical patterns have been observed to be symmetrically
determined within the Star matrix. Conclusion That some of the same numerical patterns
that exist within the genetic code should find their counterpart
within the Star of Israel should give us pause for thought. This
latter object, created from the CVs associated with the twelve
sons/tribes of Israel, and organised according to principles laid
out in the last five verses of Ezekiel’s eponymous prophetic
book, serves as an effective witness to phenomena previously
established at this website. Above all, the primacy of the number
37 cannot be understated in these proceedings. In his recent book, Is God a
Mathematician?, astrophysicist and author Mario Livio
explores the question of how it is that mathematics has evolved
over the last few thousand years to so accurately describe and
predict nearly all aspects of the physical world in which we
live. In casting his learned eye over the subject, one domain
that appears to have escaped his attention is the burgeoning
field of bioinformatics, although he does muse on the possibility
of its having a mathematical underpinning: The
astounding success of the physical sciences in discovering
mathematical laws that govern the behavior of the cosmos at
large raised the inevitable question of whether or not
similar principles might also underlie biological, social, or
economic processes. Is mathematics only the language of
nature, mathematicians wondered, or is it also the language
of human nature? (2009:122)
Another
field that we might consider adding to the discussion is that of
biblical gematria. The parallels between the gematria-based Star
of Israel and the nucleon counts of the 20 canonical amino acids
– contextualised by the dual criteria of symmetry and
‘divisibility by 037’ – offer a striking
demonstration of how rudimentary numerical phenomena appear to
permeate the very fabric of reality. For us this should occasion
no great surprise, as all are the handiwork of the single
Creator, the God of the Judeo-Christian Scriptures. Stephen
Coneglan BA 2010-06-18